Models comparison
Gabriele Pittarello
2025-06-12
Source:vignettes/modelscomparison.Rmd
modelscomparison.RmdIn this vignette, we compare the performance of the claim development
models implemented in the clmplus package with the
performance of the claim amount models available in the apc
package (Fannon and Nielsen (2020)). The
comparison is performed based on the error incidence on the reserve
(,
defined below).
The code replicates on a smaller number of datasets the analysis in
section 4.3. Benchmarking on Multiple Data Sets of the
manuscript Pittarello, Hiabu, and Villegas
(2025). We will use publicly available datasets from the R
packages clmplus, ChainLadder, and
apc for this comparison.
Let us denote with the accident period and the development period respectively. The error incidence on the reserve is defined in the manuscript as the sum of the predicted incremental payments for calendar periods larger than (the claims reserve, ) to the sum of true incremental payments for calendar periods larger than .
list.of.datasets <- list(
GenIns=GenIns,
sifa.mod=sifa.mod,
sifa.gtpl=sifa.gtpl,
sifa.mtpl=sifa.mtpl,
amases.gtpl=amases.gtpl,
amases.mod=amases.mod,
amases.mtpl=amases.mtpl,
bz = incr2cum(data.loss.BZ()$response),
ta = incr2cum(data.loss.TA()$response),
xl = incr2cum(data.loss.XL()$response),
vnj = incr2cum(data.loss.VNJ()$response),
abc=ABC,
autoC= auto$CommercialAutoPaid,
autoP = auto$PersonalAutoPaid,
autoBI = AutoBI$AutoBIPaid,
mclpaid= MCLpaid,
medmal=MedMal$MedMalPaid,
mortgage=Mortgage,
mw08=MW2008,
mw14=MW2014,
ukmotor = UKMotor,
usapaid=USAApaid
)Code for Section 4.3.1. Models Ranking
We rank the different models based on a cross-validation scheme. The training-validation split we use is represented in the following picture.
# models ranking
J=12
df<-data.frame(expand.grid(c(0:(J-1)),c(0:(J-1))),c(1:(J^2)))
colnames(df) <- c("origin","dev","value")
df$value[df$origin+df$dev==(J-1)]=c(2)
df$value[df$origin+df$dev<(J-1)]=c(1)
df$value[df$origin+df$dev>=J]=c(NA)
df[J,3]=c(NA)
df[J*J-J+1,3]=c(NA)
ggplot(data=df, aes(x=as.integer(dev), y=as.integer(origin))) +
geom_tile(aes(fill = as.factor(value),color="#000000"))+scale_y_reverse()+
scale_fill_manual(values=c("royalblue", "darkred", "white"),
na.value = "white",
labels=c("Train","Validation",""))+
theme_classic()+
labs(x="Development year", y="Accident year",fill="")+
theme(axis.title.x = element_text(size=8), axis.text.x = element_text(size=7))+
theme(axis.title.y = element_text(size=8), axis.text.y = element_text(size=7))+
scale_color_brewer(guide = 'none')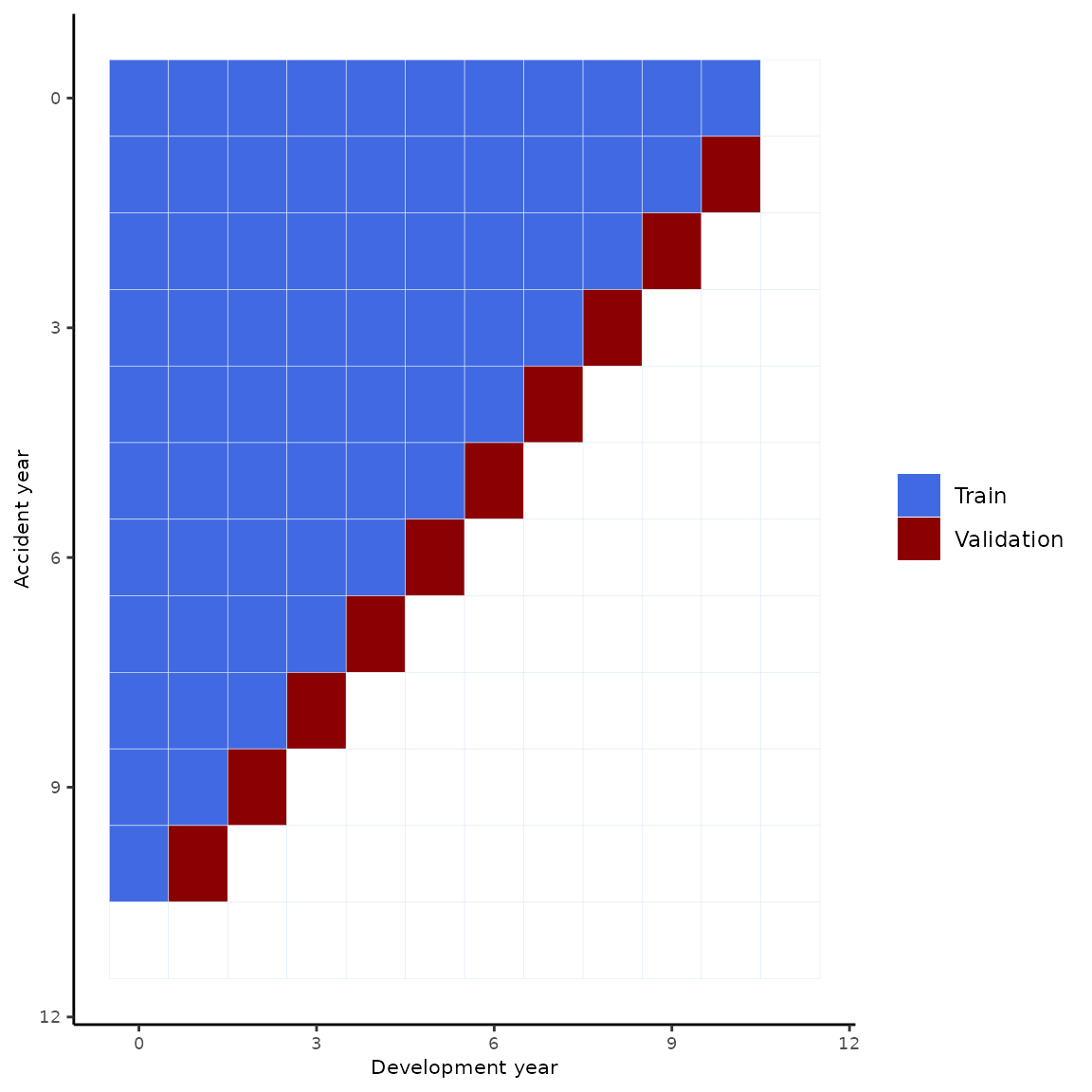
modelsranking.1d <- function(data.T){
"
Function to rank the clmplus package and apc package age-period-cohort models.
This function takes a triangle of cumulative payments as input.
It returns the ranking on the triangle.
"
leave.out=1
model.name = NULL
error.incidence = NULL
mre = NULL
#pre-processing
triangle <- data.T$cumulative.payments.triangle
J <- dim(triangle)[2]
reduced.triangle <- c2t(t2c(triangle)[1:(J-leave.out),1:(J-leave.out)])
newt.rtt <- AggregateDataPP(reduced.triangle)
to.project <- t2c(triangle)[1:(J-leave.out-1),J-leave.out]
true.values <- t2c(triangle)[2:(J-leave.out),J]
for(ix in c('a','ac','ap','apc')){
hz.fit <- StMoMo::fit(models[[ix]],
Dxt = newt.rtt$occurrance,
Ext = newt.rtt$exposure,
wxt=newt.rtt$fit.w,
iterMax=as.integer(1e+05))
hz.rate = fcst.fn(hz.fit,
hazard.model = ix,
gk.fc.model = 'a',
ckj.fc.model= 'a')$rates[,1]
fij = (2+hz.rate)/(2-hz.rate)
pred.fij = fij[(leave.out+1):length(fij)]
pred.v=to.project*pred.fij
r.errors = (pred.v-true.values)/true.values
error.inc.num = sum(pred.v-true.values,na.rm = T)
error.inc.den = sum(true.values)
model.name = c(model.name,
paste0('clmplus.',ix))
error.incidence = c(error.incidence,error.inc.num/error.inc.den)
mre = c(mre,mean(r.errors))
}
# ix='lc'
# hz.fit <- fit.lc.nr(data.T = newt.rtt,
# iter.max = 3e+04)
# if(hz.fit$converged==TRUE){hz.rate = forecast.lc.nr(hz.fit,J=dim(newt.rtt$cumulative.payments.triangle)[2])$rates[,1:leave.out]
# fij = (2+hz.rate)/(2-hz.rate)
# pred.fij = fij[(leave.out+1):length(fij)]
# pred.v=to.project*pred.fij
# r.errors = (pred.v-true.values)/true.values
#
# error.inc.num = sum(pred.v-true.values,na.rm = T)
# error.inc.den = sum(true.values)
#
# model.name = c(model.name,
# paste0('clmplus.',ix))
# error.incidence = c(error.incidence,error.inc.num/error.inc.den)
# mre = c(mre,mean(r.errors))}
out1 <- data.frame(
model.name,
# mre,
error.incidence)
## APC package
newt.apc <- apc.data.list(response=newt.rtt$incremental.payments.triangle,
data.format="CL")
## apc
rmse = NULL
mae = NULL
error.pc = NULL
model.name = NULL
error.incidence = NULL
model.family = NULL
mre = NULL
true.inc.values <- t2c(data.T$incremental.payments.triangle)[2:(J-leave.out),(J-leave.out+1):J]
for(apc.mods in c("AC","APC")){ #,"AP"
fit <- apc.fit.model(newt.apc,
model.family = "od.poisson.response",
model.design = apc.mods)
if(apc.mods == "AC"){fcst <- apc.forecast.ac(fit)$trap.response.forecast}
# if(apc.mods == "AP"){fcst <- apc.forecast.ap(fit)$trap.response.forecast}
if(apc.mods == "APC"){fcst <- apc.forecast.apc(fit)$trap.response.forecast}
plogram.hat = t2c.full.square(incr2cum(t(fcst)))
pred.v = plogram.hat[,(J-leave.out+1):J]
pred.v = pred.v[2:length(pred.v)]
r.errors = (pred.v-true.values)/true.values
error.inc.num = sum(pred.v-true.values)
error.inc.den = sum(true.values)
model.name = c(model.name,
paste0('apc.',tolower(apc.mods)))
error.incidence = c(error.incidence,error.inc.num/error.inc.den)
mre = c(mre,mean(r.errors))
}
out2 <- data.frame(
model.name,
# mre,
error.incidence)
out3 <- rbind(out1,out2)
out3 <- out3[order(abs(out3$error.incidence),decreasing = F),]
out3[,'ei.rank']=c(1:dim(out3)[1])
# out3[,'mre.rank']=order(abs(out3$mre),decreasing = F)
#fix it manually
r2set=min(out3$ei.rank[out3$model.name=='apc.ac'],
out3$ei.rank[out3$model.name=='clmplus.a'])
out3$ei.rank[out3$model.name=='apc.ac']=r2set
out3$ei.rank[out3$model.name=='clmplus.a']=r2set
if( out3$ei.rank[out3$model.name=='apc.ac'] < max(out3$ei.rank)){
cond=out3$ei.rank>out3$ei.rank[out3$model.name=='apc.ac']
out3$ei.rank[cond]=out3$ei.rank[cond]-1
}
return(list(models.ranks=out3))
}
modelsranking <- function(list.of.datasets){
"
This functions returns the datasets to plot in the ranking section of the paper.
The input is a list of datasets that constitue the sample.
The output is the rankings across different data sources.
"
full.ranks=NULL
for(df.ix in names(list.of.datasets)){
out.df=modelsranking.1d(AggregateDataPP(list.of.datasets[[df.ix]]))
out.df$models.ranks[,'data.source']=rep(df.ix,dim(out.df$models.ranks)[1])
full.ranks=rbind(full.ranks,out.df$models.ranks)
}
return(list(full.ranks=full.ranks))
}
p_min_expd0 <- ggplot(full.ranks$full.ranks, aes(model.name, data.source)) +
geom_tile(aes(fill = cut(ei.rank, breaks=0:6, labels=1:6)), colour = "grey") +
ggtitle(" ") +
theme_classic()+
geom_text(aes(label = ei.rank))+
scale_y_discrete(limits=names(list.of.datasets)) +
scale_fill_manual(drop=FALSE, values=colorRampPalette(c("white","#6699CC"))(6), na.value="#EEEEEE", name="Rank") +
xlab("Model") + ylab("Data source")
p_min_expd0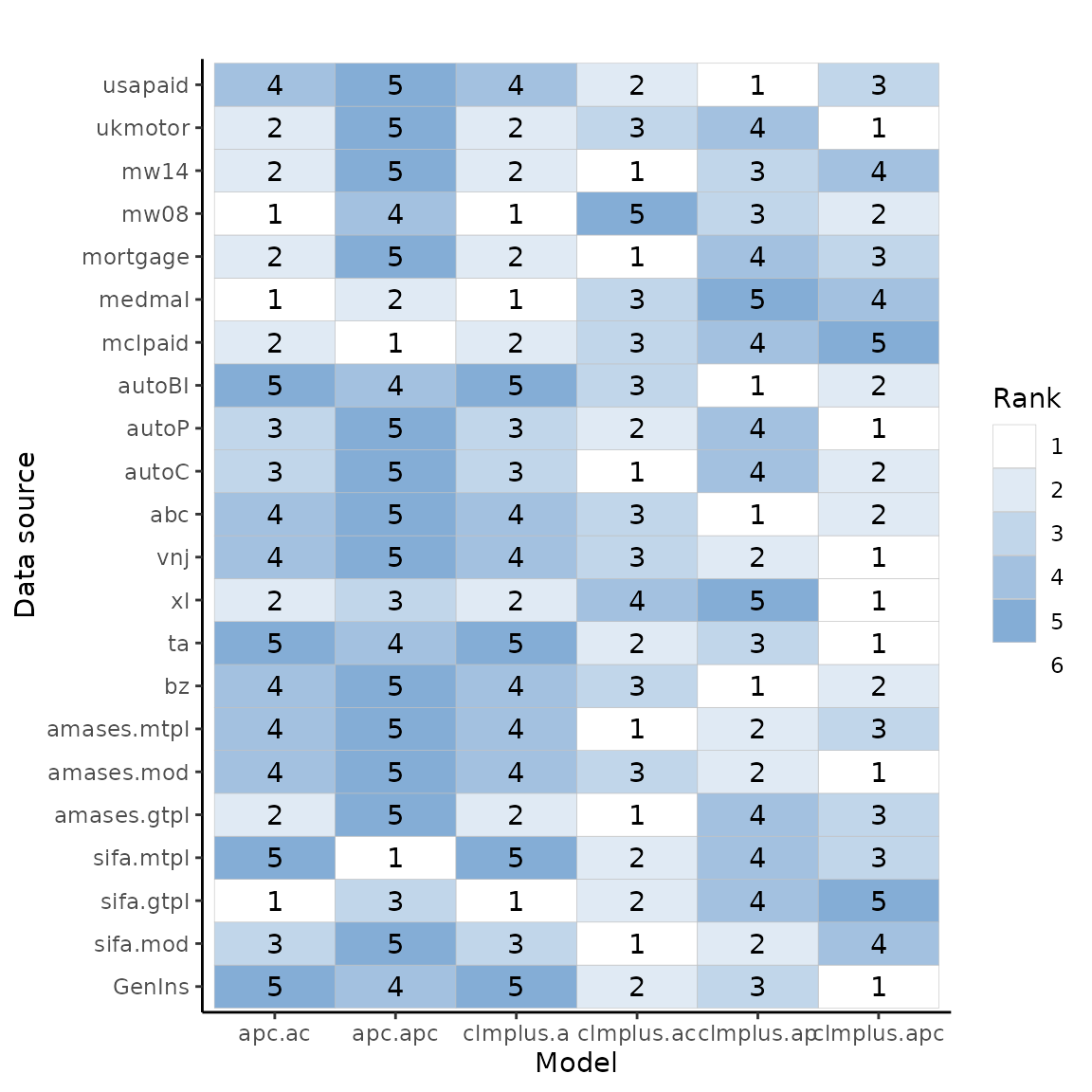
tbl=full.ranks$full.ranks %>%
dplyr::group_by(model.name) %>%
dplyr::summarise(mean.rank = mean(ei.rank))
tbl
#> # A tibble: 6 × 2
#> model.name mean.rank
#> <chr> <dbl>
#> 1 apc.ac 3.09
#> 2 apc.apc 4.14
#> 3 clmplus.a 3.09
#> 4 clmplus.ac 2.32
#> 5 clmplus.ap 3
#> 6 clmplus.apc 2.45
library(dplyr)
temp.df=full.ranks$full.ranks[,c('model.name','ei.rank')] %>%
group_by(model.name, ei.rank) %>% summarise(count = n())
#> `summarise()` has grouped output by 'model.name'. You can override using the
#> `.groups` argument.The following picture was not included in the paper but it shows the models ranks counts for each model.
ggplot(temp.df, aes(y=count, x=factor(ei.rank))) +
geom_bar(position="stack", stat="identity",fill='#6699CC') +
scale_y_continuous(limits=c(0,15))+
facet_wrap(~model.name, scales='free')+
theme_classic()+
ylab("")+
xlab("Rank")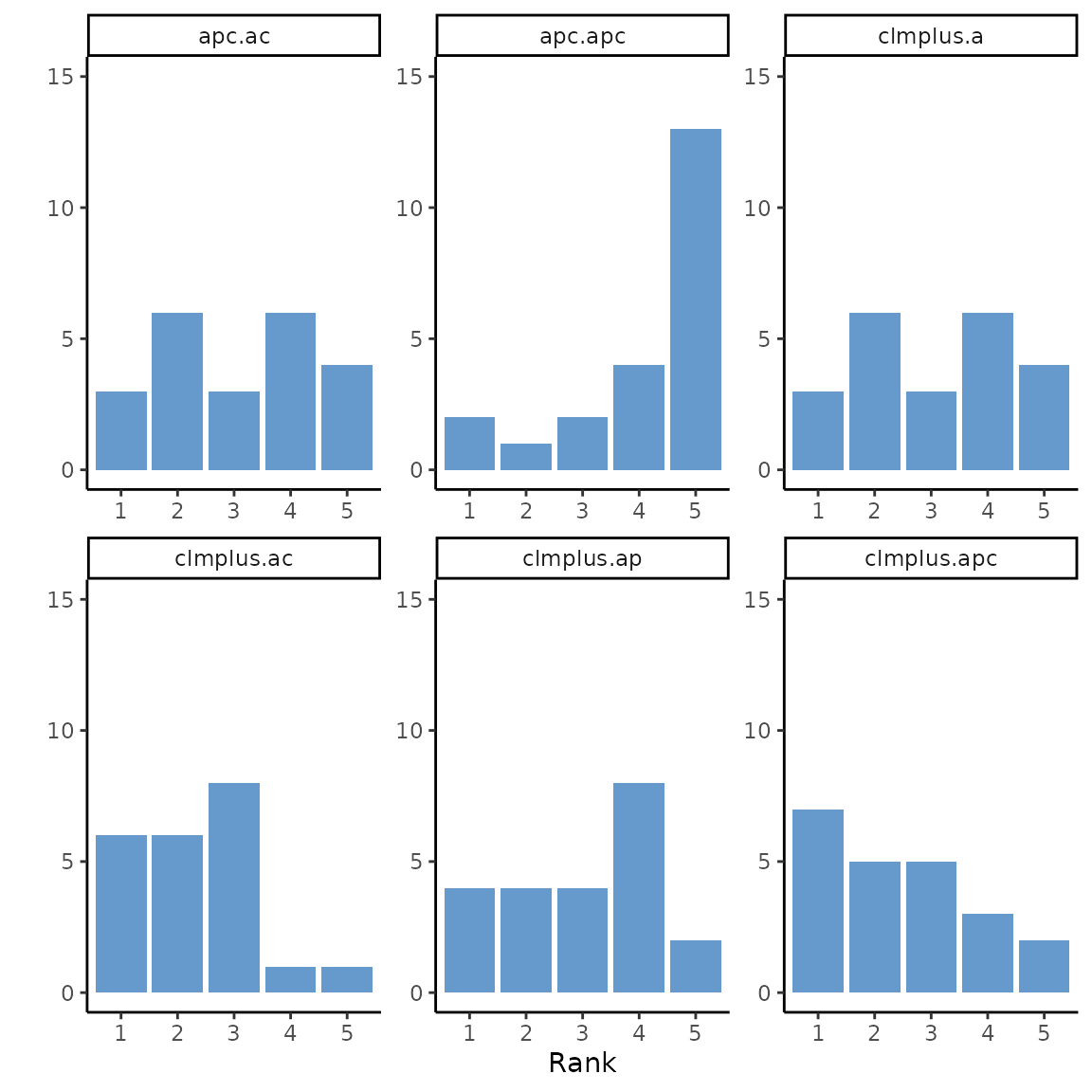
Code for Section 4.3.2. Model Families Comparison
We evaluate the out-of-sample performance of our models by using a training, validation and testing split.
J=12
df<-data.frame(expand.grid(c(0:(J-1)),c(0:(J-1))),c(1:(J^2)))
colnames(df) <- c("origin","dev","value")
df$value[df$origin+df$dev==(J-1)]=c(3)
df$value[df$origin+df$dev<(J-2)]=c(1)
df$value[df$origin+df$dev==(J-2)]=c(2)
df$value[df$origin+df$dev>=J]=c(NA)
#nas in the lower
df[J,3]=c(NA)
df[J-1,3]=c(NA)
df[J+J-1,3]=c(NA)
df[J*J-J+1,3]=c(NA)
df[J*J-J+1,3]=c(NA)
#nas in the upper tail
df[J*J-J+1-12,3]=c(NA)
df[J*J-J+2-12,3]=c(NA)
ggplot(data=df, aes(x=as.integer(dev), y=as.integer(origin))) +
geom_tile(aes(fill = as.factor(value),color="#000000"))+scale_y_reverse()+
scale_fill_manual(values=c("royalblue", "darkred", "darkgreen","white"),
na.value = "white",
labels=c("Train","Validation","Test",""))+
theme_classic()+
labs(x="Development year", y="Accident year",fill="")+
theme(axis.title.x = element_text(size=8), axis.text.x = element_text(size=7))+
theme(axis.title.y = element_text(size=8), axis.text.y = element_text(size=7))+
scale_color_brewer(guide = 'none')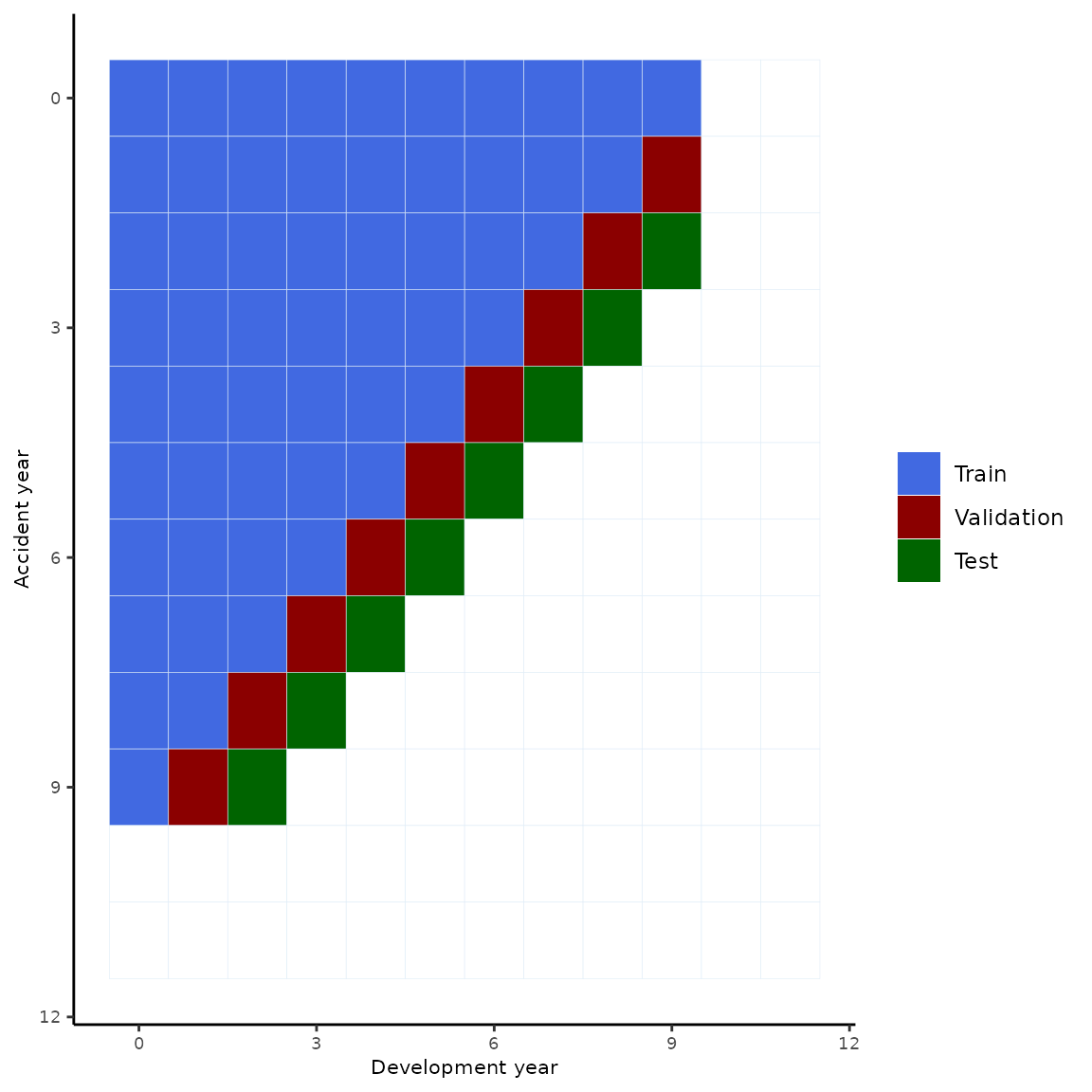
best.of.the.bests <- function(df1,df2){
"
Util to turn character columns values into numeric.
"
df1=apply(df1,MARGIN=2,FUN=as.numeric)
df2=apply(df2,MARGIN=2,FUN=as.numeric)
df3 <- rbind(df1,df2)
df3=apply(df3,FUN=abs.min,MARGIN = 2)
return(df3)
}
modelcomparison.1d <- function(cumulative.payments.triangle){
"
Function to compare the clmplus package age-period-cohort models with apc package age-period-cohort models performances across different triangles.
This function takes a triangle of cumulative payments as input.
It returns the accuracy measures for the two families on the triangle.
"
# function internal variables
leave.out=2
rmse = NULL
mae = NULL
error.pc = NULL
model.name = NULL
error.incidence = NULL
model.family = NULL
mre = NULL
# data pre-precessing ----
J <- dim(cumulative.payments.triangle)[2]
reduced.triangle <- c2t(t2c(cumulative.payments.triangle)[1:(J-leave.out),1:(J-leave.out)])
newt.rtt <- AggregateDataPP(reduced.triangle)
newt.apc <- apc.data.list(response=newt.rtt$incremental.payments.triangle,
data.format="CL")
## stmomo -----
to.project <- t2c(cumulative.payments.triangle)[1:(J-leave.out-1),J-leave.out]
true.values <- t2c(cumulative.payments.triangle)[2:(J-leave.out),(J-leave.out+1):J]
for(ix in c('a','ac','ap','apc')){ ##names(models)
hz.fit <- StMoMo::fit(models[[ix]],
Dxt = newt.rtt$occurrance,
Ext = newt.rtt$exposure,
wxt=newt.rtt$fit.w,
iterMax=as.integer(1e+05))
hz.rate = fcst.fn(hz.fit,
hazard.model = ix,
gk.fc.model = 'a',
ckj.fc.model= 'a')$rates[,1:leave.out]
J.new=dim(reduced.triangle)[2]
fij = (2+hz.rate)/(2-hz.rate)
pred.mx = fij
pred.mx[,1]=fij[,1]*c(NA,to.project)
temp=unname(pred.mx[1:(J.new-1),1][!is.na(pred.mx[1:(J.new-1),1])])
pred.mx[,2]=fij[,2]*c(rep(NA,J.new-length(temp)),temp)
true.mx= rbind(rep(NA,2),true.values)
# this is meant to be NA
true.mx[2,2]=NA
sq.errors = (pred.mx-true.mx)^2
abs.errors = abs(pred.mx-true.mx)
r.errors = (pred.mx-true.mx)/true.mx
error.inc.num = apply(pred.mx-true.mx,sum,MARGIN=2,na.rm=T)
error.inc.den = apply(true.mx,sum,MARGIN=2,na.rm=T)
model.name.ix = c(paste0(ix,".val"),paste0(ix,".test"))
model.name = c(model.name,model.name.ix)
model.family = c(model.family,rep(ix,2))
rmse = c(rmse,sqrt(apply(sq.errors,MARGIN = 2,mean,na.rm=T)))
mae = c(mae,apply(abs.errors,MARGIN = 2,mean,na.rm=T))
mre = c(mre,apply(r.errors,MARGIN = 2,mean,na.rm=T))
error.incidence = c(error.incidence,error.inc.num/error.inc.den)
}
## stmomo results ----
out1 <- data.frame(
model.name,
model.family,
mre,
error.incidence,
rmse,
mae)
temp.ix <- grepl(".val", model.name)
temp.df <- out1[temp.ix,]
out2 <- data.frame(
rmse=temp.df$model.name[which(abs(temp.df$rmse)==min(abs(temp.df$rmse)))],
mre=temp.df$model.name[which(abs(temp.df$mre)==min(abs(temp.df$mre)))],
mae=temp.df$model.name[which(abs(temp.df$mae)==min(abs(temp.df$mae)))],
error.incidence=temp.df$model.name[which(abs(temp.df$error.incidence)==min(abs(temp.df$error.incidence)))])
temp.ix <- grepl(".test", model.name)
out3 <- out1[temp.ix,]
best.df = out2
best.df[1,]=NA
out.test.min <- data.frame(
rmse=out3$model.name[which(abs(out3$rmse)==min(abs(out3$rmse)))],
mre=out3$model.name[which(abs(out3$mre)==min(abs(out3$mre)))],
mae=out3$model.name[which(abs(out3$mae)==min(abs(out3$mae)))],
error.incidence=out3$model.name[which(abs(out3$error.incidence)==min(abs(out3$error.incidence)))])
temp.mx=matrix((sub("\\..*", "", out2) == sub("\\..*", "", out.test.min)),nrow=1)
choices.mx.clmplus=matrix(sub("\\..*", "", out2),nrow=1)
agreement.frame.clmplus=data.frame(temp.mx)
choices.frame.clmplus=data.frame(choices.mx.clmplus)
colnames(agreement.frame.clmplus)=colnames(out2)
colnames(choices.frame.clmplus)=colnames(out2)
for(col.ix in colnames(out2)){
res=out1$model.family[out1$model.name == out2[1,col.ix]]
res.test = out3$model.family == res
best.df[1,col.ix] = out3[res.test,col.ix]}
families.set=c('a','apc') #'ap',
temp.ix = out3$model.family %in% families.set
comparison.df = out3[temp.ix,]
comparison.df = cbind(comparison.df,
approach=rep('clmplus',length(families.set)))
## apc ----
rmse = NULL
mae = NULL
error.pc = NULL
model.name = NULL
error.incidence = NULL
model.family = NULL
mre = NULL
true.inc.values <- t2c(cum2incr(cumulative.payments.triangle))[2:(J-leave.out),(J-leave.out+1):J]
for(apc.mods in c("AC","APC")){ #,"AP"
fit <- apc.fit.model(newt.apc,
model.family = "od.poisson.response",
model.design = apc.mods)
if(apc.mods == "AC"){fcst <- apc.forecast.ac(fit)$trap.response.forecast}
# if(apc.mods == "AP"){fcst <- apc.forecast.ap(fit)$trap.response.forecast}
if(apc.mods == "APC"){fcst <- apc.forecast.apc(fit)$trap.response.forecast}
plogram.hat = t2c.full.square(incr2cum(t(fcst)))
pred.mx = plogram.hat[,(J-leave.out+1):J]
# true.mx= rbind(rep(NA,2),true.inc.values)
# # this is meant to be NA
# true.mx[2,2]=NA
sq.errors = (pred.mx-true.mx)^2
abs.errors = abs(pred.mx-true.mx)
r.errors = (pred.mx-true.mx)/true.mx #use same benchmark
error.inc.num = apply(pred.mx-true.mx,sum,MARGIN=2,na.rm=T)
error.inc.den = apply(true.mx,sum,MARGIN=2,na.rm=T) #use same benchmark
model.name.ix = c(paste0(apc.mods,".val"),paste0(apc.mods,".test"))
model.name = c(model.name,tolower(model.name.ix))
model.family = c(model.family,tolower(rep(apc.mods,2)))
rmse = c(rmse,sqrt(apply(sq.errors,MARGIN = 2,mean,na.rm=T)))
mae = c(mae,apply(abs.errors,MARGIN = 2,mean,na.rm=T))
mre = c(mre,apply(r.errors,MARGIN = 2,mean,na.rm=T))
error.incidence = c(error.incidence,error.inc.num/error.inc.den)}
out4 <- data.frame(
model.name,
model.family,
mre,
error.incidence,
rmse,
mae)
temp.ix <- grepl(".val", model.name)
temp.df <- out4[temp.ix,]
out5 <- data.frame(
rmse=temp.df$model.name[which(abs(temp.df$rmse)==min(abs(temp.df$rmse)))],
mre=temp.df$model.name[which(abs(temp.df$mre)==min(abs(temp.df$mre)))],
mae=temp.df$model.name[which(abs(temp.df$mae)==min(abs(temp.df$mae)))],
error.incidence=temp.df$model.name[which(abs(temp.df$error.incidence)==min(abs(temp.df$error.incidence)))])
temp.ix <- grepl(".test", model.name)
out6 <- out4[temp.ix,]
out.test.min2 <- data.frame(
rmse=out6$model.name[which(abs(out6$rmse)==min(abs(out6$rmse)))],
mre=out6$model.name[which(abs(out6$mre)==min(abs(out6$mre)))],
mae=out6$model.name[which(abs(out6$mae)==min(abs(out6$mae)))],
error.incidence=out6$model.name[which(abs(out6$error.incidence)==min(abs(out6$error.incidence)))])
temp.mx=matrix((sub("\\..*", "", out5) == sub("\\..*", "", out.test.min2)),nrow=1)
choices.mx.apc=matrix(sub("\\..*", "", out5),nrow=1)
choices.frame.apc=data.frame(choices.mx.apc)
agreement.frame.apc=data.frame(temp.mx)
colnames(agreement.frame.apc)=colnames(out5)
colnames(choices.frame.apc)=colnames(out5)
best.df.apc = out5
best.df.apc[1,]=NA
for(col.ix in colnames(out5)){
res=out4$model.family[out4$model.name == out5[1,col.ix]]
res.test = out6$model.family == res
best.df.apc[1,col.ix] = out6[res.test,col.ix]}
families.set=c('ac','apc') #'ap',
temp.ix = out6$model.family %in% families.set
comparison.df.apc = out6[temp.ix,]
comparison.df.apc = cbind(comparison.df.apc,
approach=rep('apc',length(families.set)))
out = list(
best.model.clmplus = best.df,
best.model.apc = best.df.apc,
agreement.frame.clmplus=agreement.frame.clmplus,
agreement.frame.apc=agreement.frame.apc,
choices.frame.clmplus=choices.frame.clmplus,
choices.frame.apc=choices.frame.apc,
comparison.df = rbind(comparison.df,
comparison.df.apc))
return(out)}
modelcomparison<-function(list.of.datasets){
"This functions returns the datasets to plot the bake-off section of the paper.
The input is a list of datasets that constitue the sample.
The output is datasets that contain accuracy measures.
"
best.fit=NULL
families.fit=NULL
agreement.clmplus=NULL
agreement.apc=NULL
choices.clmplus=NULL
choices.apc=NULL
for(df.ix in names(list.of.datasets)){
cat(paste0(".. Comparison on dataset: ",df.ix))
out.ix = modelcomparison.1d(list.of.datasets[[df.ix]])
best.of.the.bests.df=best.of.the.bests(out.ix$best.model.clmplus,
out.ix$best.model.apc)
out.ix$best.model.clmplus['package']= 'clmplus'
out.ix$best.model.apc['package']= 'apc'
best.of.the.bests.df['package']='overall.best'
best.fit=rbind(best.fit,
out.ix$best.model.clmplus,
out.ix$best.model.apc,
best.of.the.bests.df)
families.fit=rbind(families.fit,
out.ix$comparison.df)
agreement.clmplus=rbind(agreement.clmplus,
out.ix$agreement.frame.clmplus)
agreement.apc=rbind(agreement.apc,
out.ix$agreement.frame.apc)
choices.clmplus=rbind(choices.clmplus,
out.ix$choices.frame.clmplus)
choices.apc=rbind(choices.apc,
out.ix$choices.frame.apc)
}
best.fit[,1:4]=apply(best.fit[,1:4],MARGIN = 2,FUN = as.numeric)
families.fit[,c('mre',
'error.incidence',
'rmse',
'mae')]=apply(
families.fit[,c('mre',
'error.incidence',
'rmse',
'mae')],
MARGIN = 2,
FUN = as.numeric)
out = list(best.fit=best.fit,
families.fit=families.fit,
agreement.clmplus=agreement.clmplus,
agreement.apc=agreement.apc,
choices.clmplus=choices.clmplus,
choices.apc=choices.apc)
return(out)
}
bake.off <- function(models.comparison){
"
This function plots out the results from the previous computations.
It takes as input the resulting dataframes of model.comparison.
The output is the boxplots of the paper's bake-off section.
"
# browser()
p1<- models.comparison$best.fit[,c("rmse","mae","package")] %>%
tidyr::pivot_longer(-c(package)) %>%
ggplot(aes(x=package,y=value))+
geom_boxplot()+
facet_wrap(.~name,nrow = 1,strip.position = 'bottom')+
theme_bw()+
theme(strip.placement = 'outside',strip.background = element_blank())
p2<- models.comparison$best.fit[,c("mre","error.incidence","package")] %>%
tidyr::pivot_longer(-c(package)) %>%
ggplot(aes(x=package,y=value))+
geom_boxplot()+
facet_wrap(.~name,nrow = 1,strip.position = 'bottom')+
theme_bw()+
theme(strip.placement = 'outside',strip.background = element_blank())
abs.best=models.comparison$best.fit[,c("mre","error.incidence","package")]
abs.best[,c("mre","error.incidence")]=apply(abs.best[,c("mre","error.incidence")],
MARGIN=2,
FUN=abs)
p3<- abs.best %>%
tidyr::pivot_longer(-c(package)) %>%
ggplot(aes(x=package,y=value))+
geom_boxplot()+
facet_wrap(.~name,nrow = 1,strip.position = 'bottom')+
theme_bw()+
theme(strip.placement = 'outside',strip.background = element_blank())
only.ei=models.comparison$best.fit[,c("error.incidence","package")]
only.ei[,c("error.incidence")]=abs(only.ei[,c("error.incidence")])
p4<- abs.best %>%
tidyr::pivot_longer(-c(package)) %>%
ggplot(aes(x=package,y=value))+
geom_boxplot()+
# facet_wrap(.~name,nrow = 1,strip.position = 'bottom')+
theme_bw()+
theme(strip.placement = 'outside',strip.background = element_blank())
out = list(p1=p1,
p2=p2,
p3=p3,
p4=p4)
return(out)
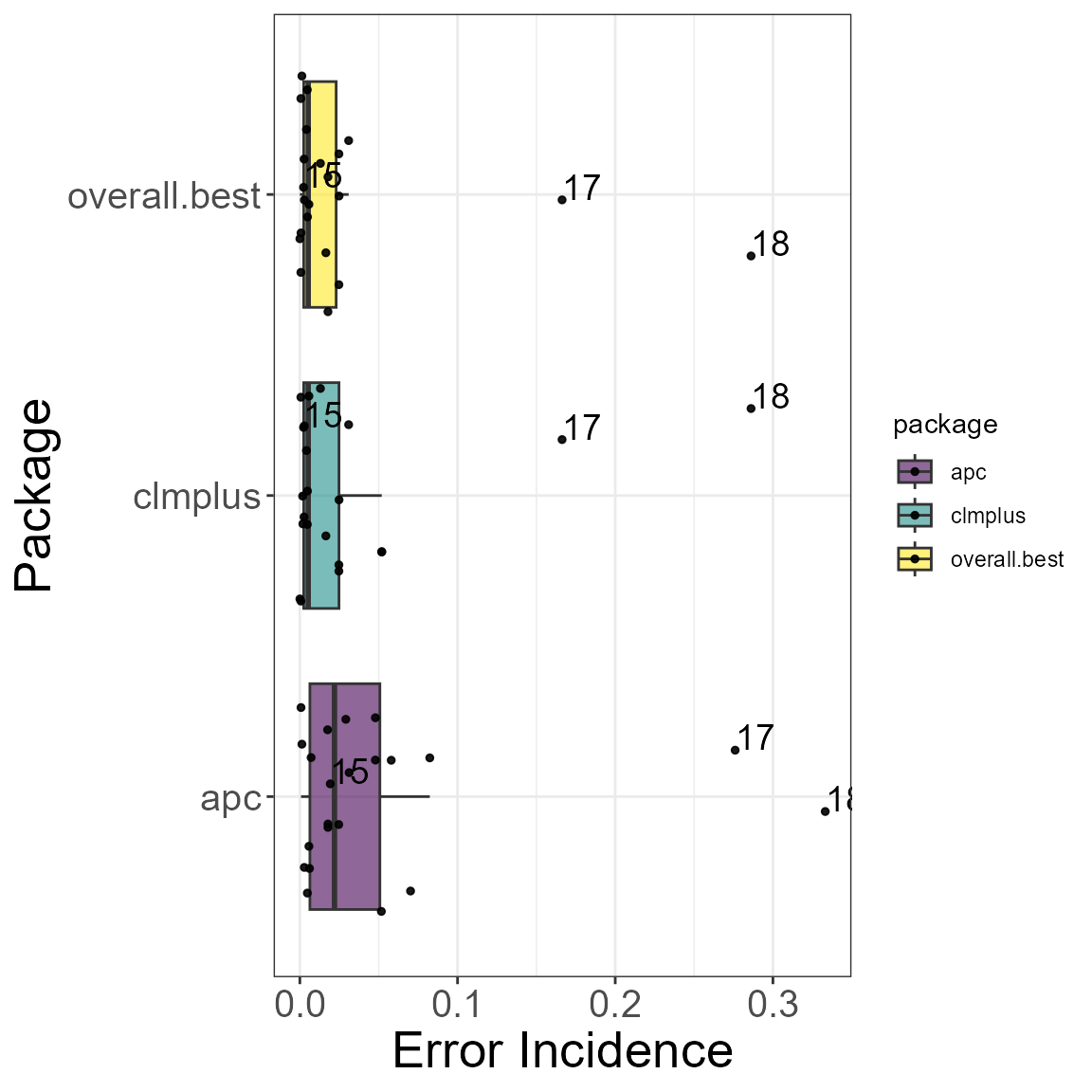
}The models in the clmplus package are compared to those
in the apc package. Below it can be found the code we used
to create the box-plot in figure 8 of our paper.
For each case we are able to pick the best model based on the error incidence.